
De novo mutations (DNMs) arise spontaneously in germline cells (de novo germline mutation) or shortly after fertilization (post-zygotic mutation). They represent the most extreme form of rare genetic mutations that have been proved as important contributors to sporadic genetic diseases, such as autism spectrum disorders, intellectual disability, epileptic encephalopathy, schizophrenia, congenital heart disease, type 1 diabetes, and hearing loss. However, not all DNMs are the causes of sporadic diseases. It is indicated that averagely 74 de novo SNVs and 3 de novo INDELs arise spontaneously in an individual's genome and only a very few of them are considered to be pathogenic. Therefore, it is a great challenge to accurately assess the causality of DNMs as well as identify disease-causative genes from the considerable number of DNMs occurred in proband. A common method to this problem is to identify genes that harbor significantly DNMs than expected by chance. However, this method requires expected background de novo mutation rates (DNMRs) of each individual gene or large normal samples as controls. If accurate background DNMRs for each gene were provided, we could identify candidate genes much easier with strong statistical support and not require normal samples as controls.
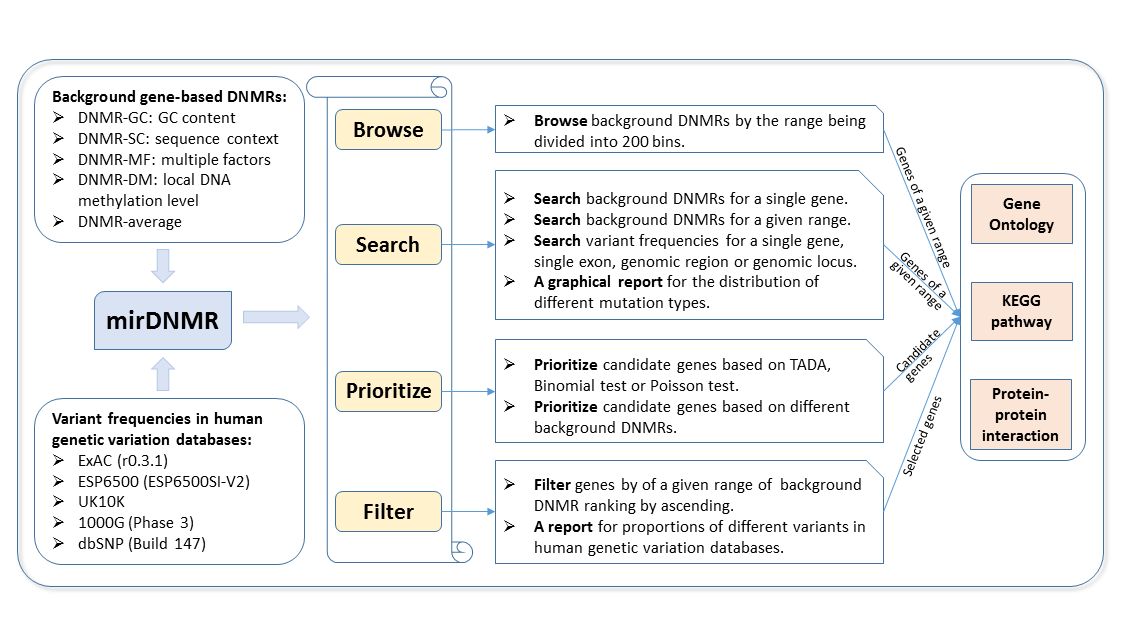
Here we constructed a gene-centered database named mirDNMR for collection of background DNMRs and variant frequencies in human genetic variation databases including ExAC (r0.3.1), ESP6500 (ESP6500SI-V2), UK10K, the 1000 Genomes (Phase 3) and dbSNP (Build 147). Users can freely browse and search resources in this database. Meanwhile, mirDNMR provides two convenient tools for users to prioritize and filter candidate genes.
Source of background DNMRs:
DNMR by GC content (DNMR-GC): Given that the DNMRs of GC bases are 1.76-fold greater than that of AT bases, it could be inferred that GC content could explain the variation of DNMRs to a large extent. DNMRs were calculated based on the whole exome sequencing data of the 200 quartets considering both gene sizes and GC contents. (Sanders et al. Nature, 2012)
DNMR by sequence context (DNMR-SC): It is known that sequence context is also an important factor that influence DNMR, so Samocha et al. created a mutation rate matrix to determine the probability for each type of tri-nucleotide variations. Based on this matrix, background DNMRs for each gene were calculated. (Samocha et al. Nature Genetics, 2014)
DNMR by multiple factors (DNMR-MF): Multiple factors influence the occurrence of DNM, such as gene length, mutation type, sequence context, biological evolution and recombination rate. Therefore, Francioli et al. build a complex model combining all these factors to assess the mutability of each locus in the human genome. Based on this model, background DNMRs for each gene were calculated. (Francioli et al. Nature Genetics, 2015)
DNMR by local DNA methylation level (DNMR-DM): Because spontaneous deamination of 5-methylcytosine results in about 14-fold higher C>T substitution rates as compared to the genome-wide average, it could be inferred that DNA methylation increase the mutability of C>T base substitution. By deep research in the relationship between human sperm local DNA methylation level and DNMR, we built a model and predicted DNMRs for each gene.
Currently available human genetic variation databases:
ExAC: The Exome Aggregation Consortium (ExAC, version r0.3.1) contains a wide variety of large-scale sequencing data spans 60,706 unrelated individuals, including African American, African American, East Asian, Finnish, Non-Finnish European, South Asian and Others.
ESP6500: The NHLBI GO Exome Sequencing Project (ESP) performed exome sequencing for a large samples. The current EVS data release (ESP6500SI-V2) is taken from 6,503 samples including European Americans and African Americans.
UK10K: The UK10K project identified rare genetic variants in 10,000 samples, including 4,000 whole genome cohorts, 3,000 neurodevelopment sample sets, 2,000 obesity sample sets and 1,000 rare diseases sample sets.
The 1000 Genomes: The 1000 Genomes Project (Phase 3) performed whole genome sequencing of 2,504 individuals from 26 different populations.
dbSNP: Build 147 (only germline) was incorporated in mirDNMR.
Browse background gene-based DNMRs (DNMR-GC, DNMR-SC, DNMR-MF, DNMR-DM and DNMR-average) based on the range of DNMR being split into 200 bins.
Search background gene-based DNMRs and variant frequencies in human genetic variation databases (ExAC, ESP6500, UK10K, the 1000 Genomes, dbSNP) for single gene, single exon, genomic region or locus (GRCH37/hg19).
Prioritize candidate genes based on DNM burden by comparing with background DNMRs using TADA, Binomial test or Poisson test.
Filter gene list based on background DNMRs and distribution of different types of variants in human genetic variation databases.
Download background gene-based DNMRs (DNMR-GC, DNMR-SC, DNMR-MF, DNMR-DM and DNMR-average) and DNM list for intellectual disability and control from trio-based WES/WGS.
This database is free and open to all users and there is no login requirement !
Jiang, Y., Li, Z., Liu, Z., Chen, D., Wu, W., Du, Y., Ji, L., Jin, Z.B., Li, W. and Wu, J. (2017) mirDNMR: a gene-centered database of background de novo mutation rates in human. Nucleic acids research, 45, D796-D803.